
|
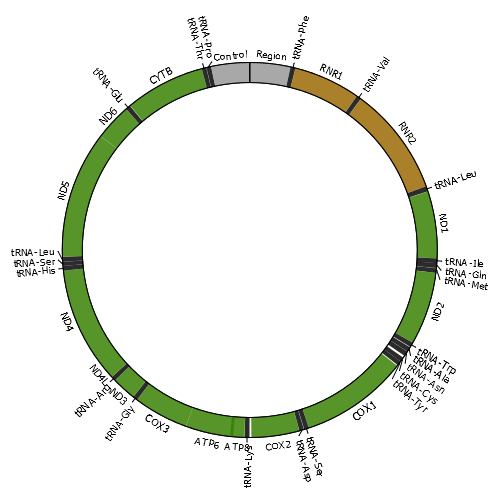
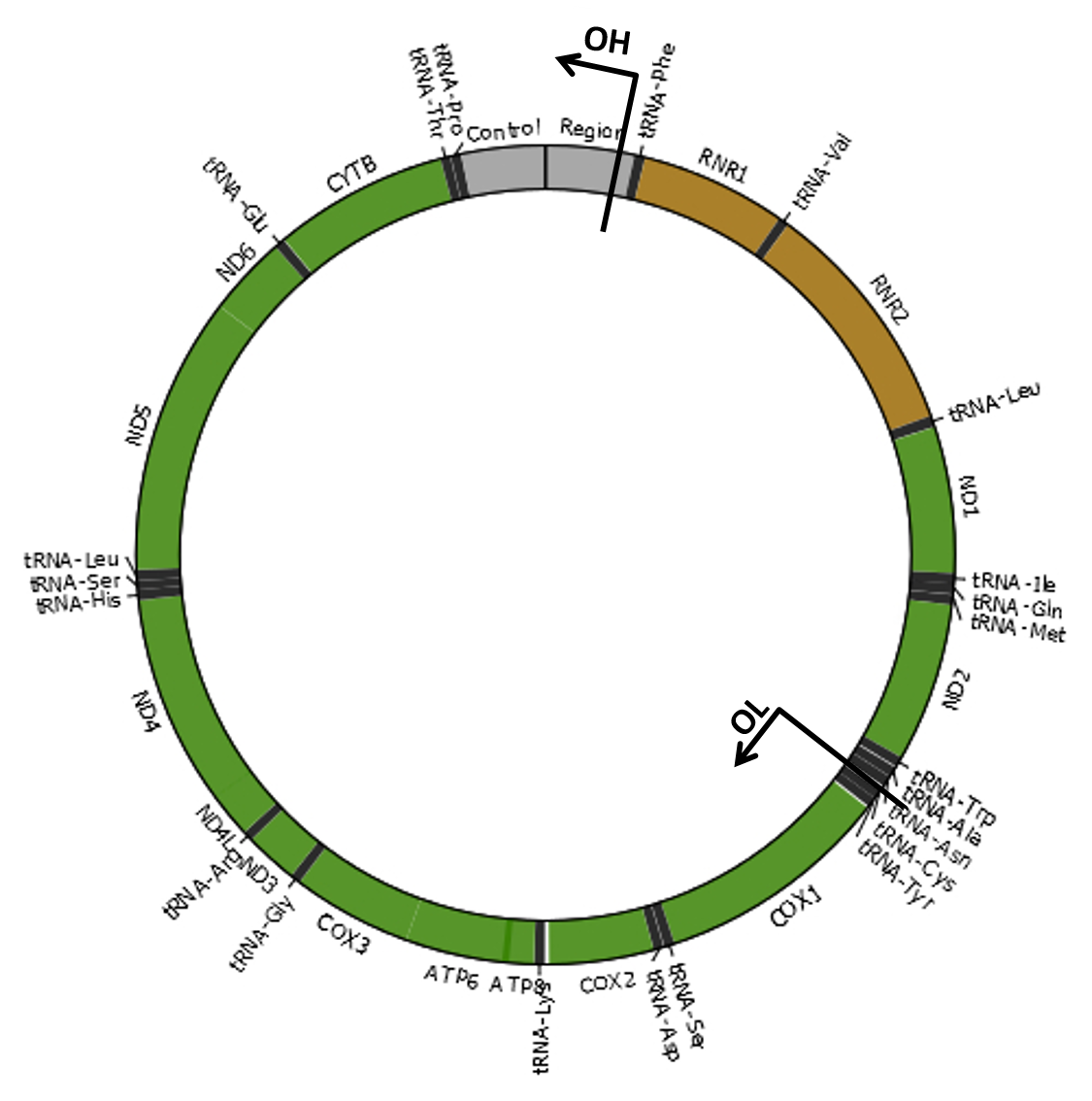
Mitochondria are cellular organelles that provide most of the energy that organisms need to live. These semi-autonomous organelles possess their own genetic material (mitochondrial DNA or mtDNA), which is necessary for the formation of essential proteins of the oxidative phosphorylation (OXPHOS) system. The mitochondrial genome of most species consists of a single circular double-stranded DNA molecule. For example, the human mtDNA is organized as shown in the image. Damage to the mitochondrial genome might occur in the form of point mutations, large deletions or duplications and DNA breakage with subsequent linearization of the mtDNA molecule. As eukaryotic cells contain many copies of mtDNA, a mutated type of mtDNA must first reach a threshold level by clonally expanding within a cell before it can cause adverse effects. The accumulation of damaged mtDNA molecules in tissues is an important cause of mitochondrial disease, a clinically heterogeneous group of disorders related with OXPHOS dysfunction. Moreover, mutated mtDNAs are suspected to contribute to the etiology of a number of age-related disorders, by accumulating with age in a variety of tissues. |
|
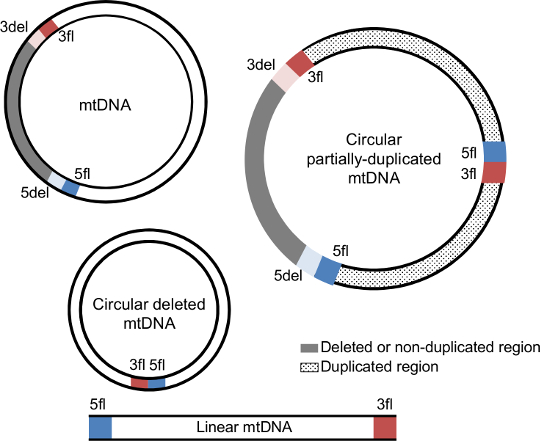
MitoBreak has information for the three most common types of mtDNA rearrangements:
|
|
Breakpoints are locations in the mtDNA where breaks have occurred. The breakpoints available in this database were always numbered according to the positions of reference mtDNA sequences (which are usually the first to be sequenced).
| Species | Reference sequence |
|---|---|
| Homo sapiens | NC_012920.1 |
| Mus musculus | NC_005089.1 |
| Rattus norvegicus | NC_001665.2 |
| Macaca mulatta | NC_005943.1 |
| Drosophila melanogaster | NC_001709.1 |
| Caenorhabditis elegans | NC_001328.1 |
| Podospora anserina | NC_001329.3 |
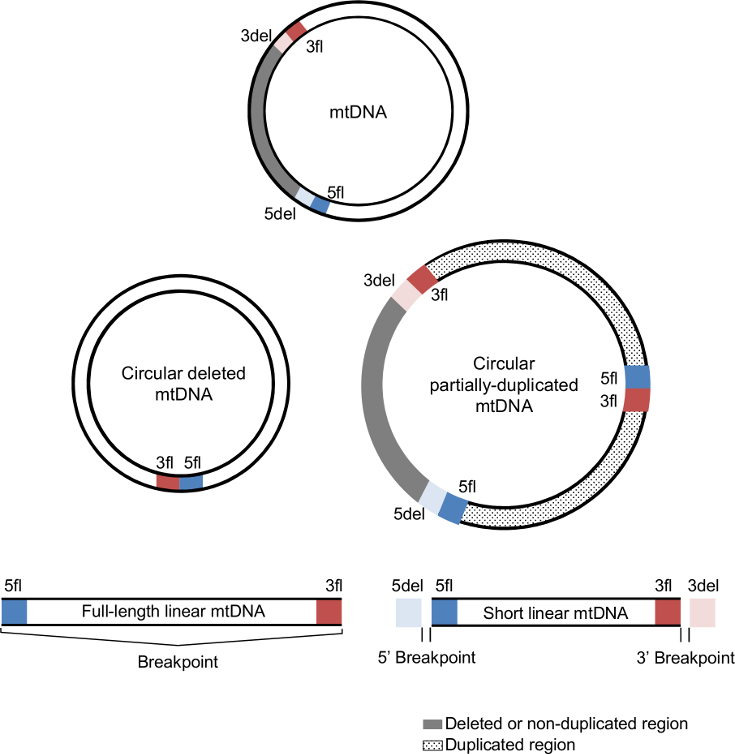
Each mtDNA deletion and duplication is defined by a unique combination of two breakpoints (5' and 3'). The 5' breakpoints are upstream of the 5' break and 3' breakpoints are downstream of the 3' break using the reference numbering. In the case of mtDNA deletions, breakpoints represent the mtDNA positions that are retained in the deleted mtDNA molecule and that flank the deleted region.

|
The deleted mtDNA molecule retains the upstream region of the 5' breakpoint (5fl; blue section) and the downstream region of the 3' breakpoint (3fl; red section) next to each other, while the removed segment (grey section) is delimitated by the downstream region of the 5' breakpoint (5del; light blue) and the upstream region of the 3' breakpoint (3del; light red). 5del and 3del also denote the flanking region of the non-duplicated region in mtDNA duplications. In addition to these four regions, duplications also have a junction site connecting 5fl and 3fl. |
|
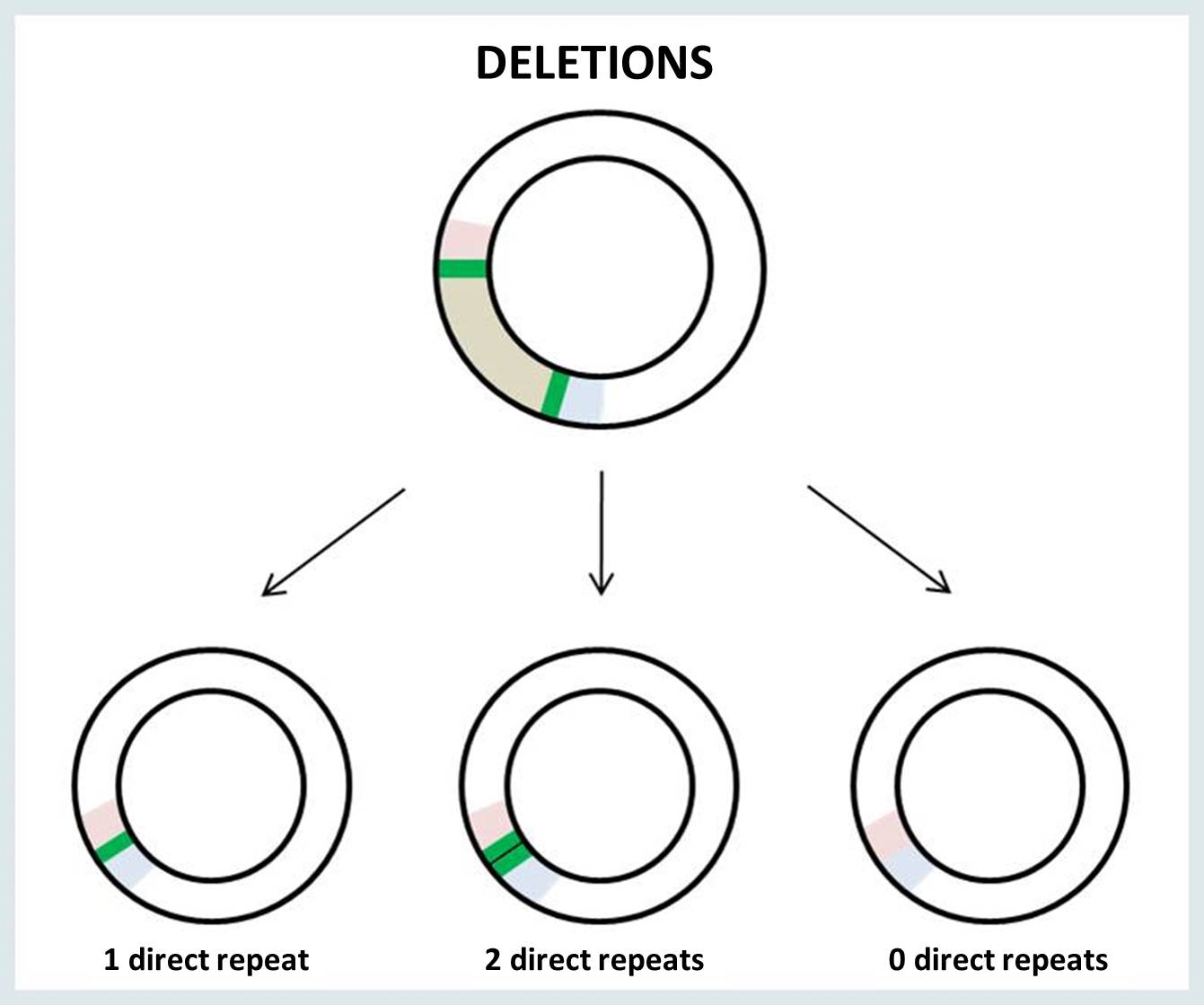
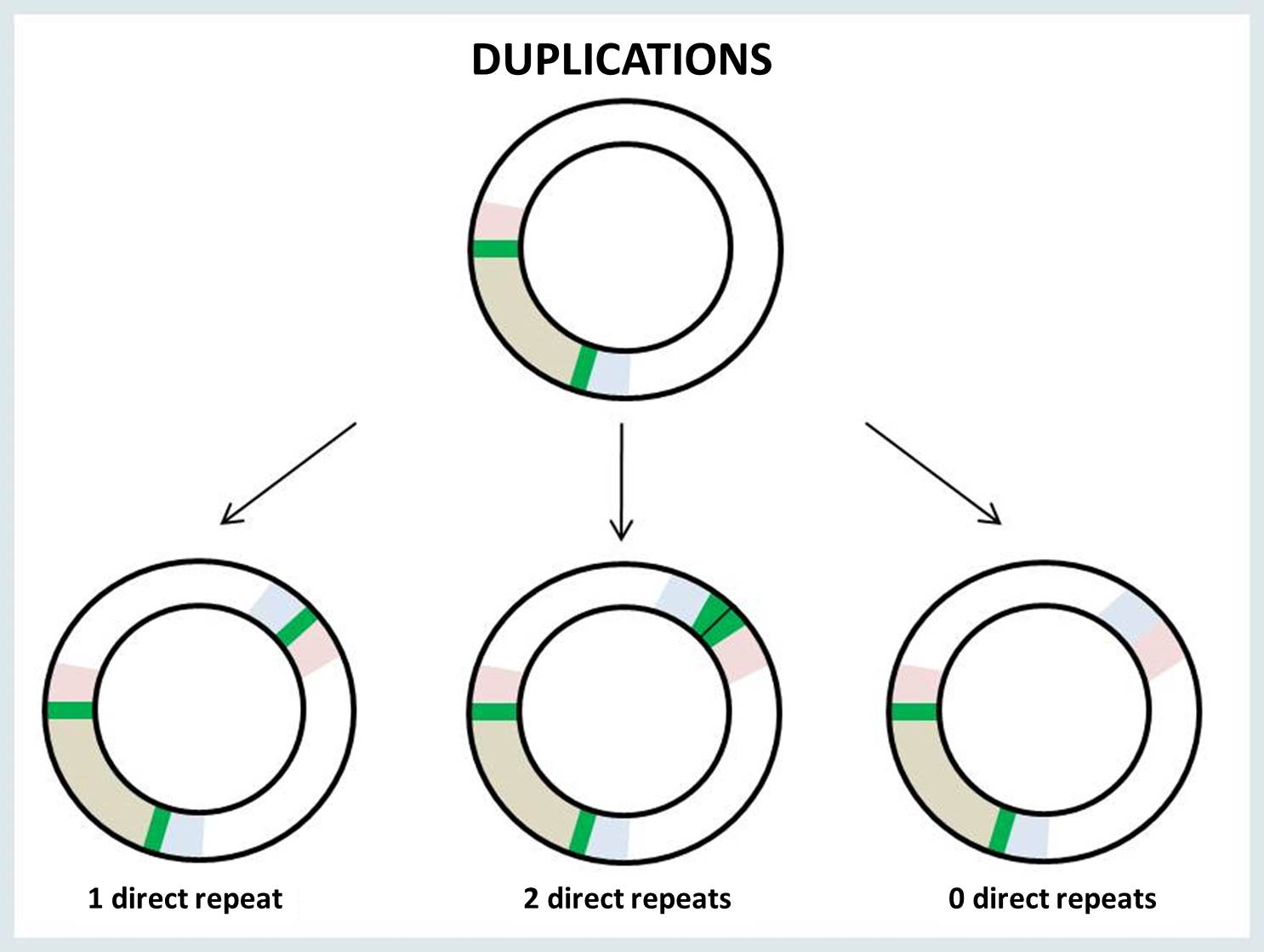
The breakpoints of mtDNA rearrangement are often located within perfect direct repeats. In such cases, there are three possible outcomes of the rearrangement: the deletion or duplication junction may retain 0, 1 or 2 copies of the direct repeat.
|
|
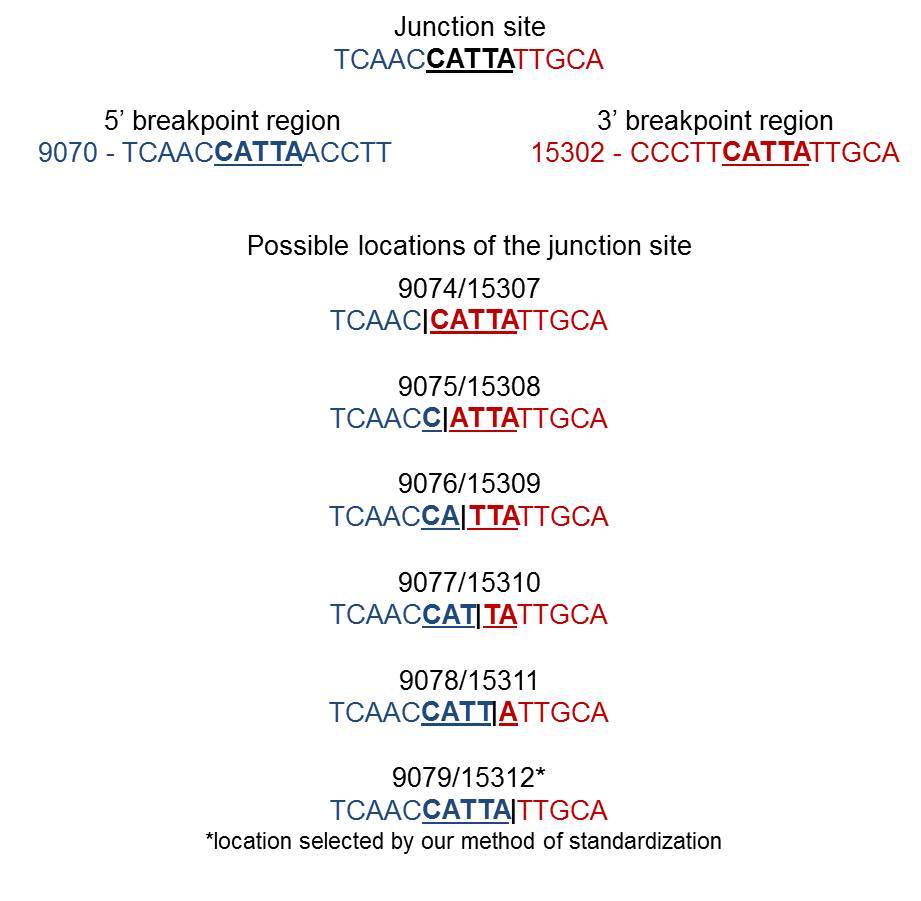
When a single copy of the direct repeat is retained in the rearrangement junction, it is impossible to identify the exact sites of mtDNA breakage (i.e., 5’ and 3’ breakpoints). There is no way to know for sure which of the two direct repeats are retained in the surrounding bases at the junction site. The following example illustrates the possible locations of the junction site in the presence of a perfect direct repeat (bold underlined nucleotides) at 5’ and 3’ breakpoints:
|
By this reason, the same rearrangement is sometimes described in the literature by arbitrary breakpoint positions inside the homologous region or by an interval of values (e.g., 9074–9079:15307–15312). All breakpoints available in MitoBreak have been assigned to the 3’ nucleotide of the direct repeat for standardization purposes (in the previous example, 9079:15312). In other words, breakpoints were always placed downstream (on the right of) the homology.
The 'Classifier tool' can be used to verify if breakpoints are located in direct repeats. It also presents the correct breakpoint numbering according to the reference mtDNA and our method of standardization.
|
Unless stated otherwise, we always refer to the origins of H-strand (OH) and L-strand (OL) replication using the strand-displacement model of mtDNA replication (Robberson et al., 1972; Berk and Clayton, 1974). Although each replication origin is a range of nucleotides, we used here the nucleotide positions 407 and 5747 for OH and OL, respectively (Chang and Clayton, 1985; Hixson et al., 1986), which included the mtDNA regions encoding the RNA fragment used as primers to initiate DNA synthesis. Therefore, the minor arc (shortest mtDNA section between OL and OH) is located between positions 408 and 5746 and the major arc (largest mtDNA section between OL and OH) is between positions 5747 and 407. References Robberson DL, Kasamatsu H, Vinograd J. 1972. Replication of mitochondrial DNA. Circular replicative intermediates in mouse L cells. Proc Natl Acad Sci U S A 69:737-741. Berk AJ, Clayton DA. 1974. Mechanism of mitochondrial DNA replication in mouse L-cells: asynchronous replication of strands, segregation of circular daughter molecules, aspects of topology and turnover of an initiation sequence. J Mol Biol 86:801-824. Chang DD, Clayton DA. 1985. Priming of human mitochondrial DNA replication occurs at the light-strand promoter. Proc Natl Acad Sci U S A 82:351-355. Hixson JE, Wong TW, Clayton DA. 1986. Both the conserved stem-loop and divergent 5'-flanking sequences are required for initiation at the human mitochondrial origin of light-strand DNA replication. J Biol Chem 261:2384-2390. |
|
The following abbreviations are used in the MitoBreak database:
General Disclaimer
Although substantial effort has been made to provide accurate and complete information, neither the authors nor the University of Porto guarantee that there will be no errors.
If you find any error, please e-mail us (mitobreak@gmail.com). The database is restricted to academic investigators. Commercial entities are required to submit a request for commercial license.